Services on Demand
article
Indicators
Share
Journal of Human Growth and Development
Print version ISSN 0104-1282On-line version ISSN 2175-3598
J. Hum. Growth Dev. vol.32 no.2 Santo André May/Aug. 2022
http://dx.doi.org/10.36311/jhgd.v32.13324
ORIGINAL ARTICLE
An interpretable machine learning model for COVID-19 screening
Gustavo Carreiro PinascoI; Eduardo Moreno Júdice de Mattos FarinaII; Fabiano Novaes Barcellos FilhoIII; Willer França FiorottiIII; Matheus Coradini Mariano Ferreira; Sheila Cristina de Souza CruzIV; Andre Louzada ColodetteIII; Luciene Rossati LoureiroIV; Tatiane ComérioIV; Dilzilene Cunha Sivirino FariasIV; Eliane de Fátima Almeida LimaI; Katia Valéria ManhambusqueI
IUniversidade Federal do Espírito Santo - UFES, Brazil
IIUniversidade Federal de São Paulo - UNIFESP, Brazil
IIIEscola Superior de Ciências da Santa Casa de Misericórdia de Vitória - EMESCAM, Brazil
IVPrefeitura Municipal de Vitória, Brazil
ABSTRACT
INTRODUCTION: the Coronavirus Disease 2019 (COVID-19) is a viral disease which has been declared a pandemic by the WHO. Diagnostic tests are expensive and are not always available. Researches using machine learning (ML) approach for diagnosing SARS-CoV-2 infection have been proposed in the literature to reduce cost and allow better control of the pandemic
OBJECTIVE: we aim to develop a machine learning model to predict if a patient has COVID-19 with epidemiological data and clinical features
METHODS: we used six ML algorithms for COVID-19 screening through diagnostic prediction and did an interpretative analysis using SHAP models and feature importances
RESULTS: our best model was XGBoost (XGB) which obtained an area under the ROC curve of 0.752, a sensitivity of 90%, a specificity of 40%, a positive predictive value (PPV) of 42.16%, and a negative predictive value (NPV) of 91.0%. The best predictors were fever, cough, history of international travel less than 14 days ago, male gender, and nasal congestion, respectively
CONCLUSION: we conclude that ML is an important tool for screening with high sensitivity, compared to rapid tests, and can be used to empower clinical precision in COVID-19, a disease in which symptoms are very unspecific
Keywords: COVID-19, machine learning, artificial intelligence, pandemia.
RESUMO
INTRODUÇÃO: a Doença do Coronavírus 2019 (COVID-19) é uma doença viral que foi declarada uma pandemia pela OMS. Testes diagnósticos são caros e nem sempre estão disponíveis. Pesquisas utilizando a abordagem de aprendizado de máquina (ML) para o diagnóstico de infecção por SARS-CoV-2 têm sido propostas na literatura para reduzir custos e permitir melhor controle da pandemia
OBJETIVO: nosso objetivo é desenvolver um modelo de aprendizado de máquina para prever se um paciente tem COVID-19 com dados epidemiológicos e características clínicas
MÉTODO: usamos seis algoritmos de ML para triagem de COVID-19 por meio de predição diagnóstica e fizemos uma análise interpretativa usando modelos SHAP e importâncias de recursos
RESULTADOS: nosso melhor modelo foi o XGBoost (XGB) que obteve área sob a curva ROC de 0,752, sensibilidade de 90%, especificidade de 40%, valor preditivo positivo (VPP) de 42,16% e valor preditivo negativo ( VPL) de 91,0%. Os melhores preditores foram febre, tosse, história de viagem internacional há menos de 14 dias, sexo masculino e congestão nasal, respectivamente
CONCLUSÃO: Concluímos que o ML é uma importante ferramenta de triagem com alta sensibilidade, em comparação aos testes rápidos, e pode ser usado para potencializar a precisão clínica na COVID-19, doença em que os sintomas são muito inespecíficos
Palavras-chave: COVID-19, aprendizado de máquina, inteligência artificial, pandemia.
Authors summary
Why was this study done?
This study was developed for tests of machine learning algorithms performing as a predictive tool for detecting COVID-19 infection and to analyze its approach in actual world pandemia.
What did the researchers do and find?
The researchers started the study from a collection of clinical data from a municipality at the beginning of the pandemic. Then, they trained different machine learning models to identify, from clinical signs and symptoms, which patients had COVID-19.
What do these findings mean?
Machine learning models can perform with high sensitivity, even with simple clinical characteristics. It can be useful for initial pandemics for screening and has a very low cost for its use.
INTRODUCTION
The Coronavirus Disease 2019 (COVID-19) is a viral disease caused by SARS-CoV-2 virus, which has been declared a pandemic by the WHO and already has more than 3 million infected worldwide and 230 thousand deaths1. The diagnostic tests using RT - qPCR still have doubts about their performance and reliability2 and are not always available.
The publication of articles related to using Machine Learning (ML) to support clinical decisions, classification of CT scans, and diagnosis of COVID-19 are growing and showing promising results to better deal with the pandemic3,4. There are few proposals for machine learning models diagnosing or predicting death by COVID-19 with good results using laboratory test features5,6, but none performed before the propaedeutic approach.
The objective of the present study is to create ML models to diagnose COVID-19 using clinical and demographic variables collected from medical records of patients suspected of having COVID-19.
METHODS
Study design and participants
The study sample was obtained retrospectively and cross-sectionally7 through the collection of data from hospitals, primary healthcare units, and emergency care centers that notified their suspected and confirmed cases of COVID-19 to the Health Department of Vitória "(PMV)": capital of Espírito Santo State, Brazil, with around 370.000 population. The screening was done through clinical-epidemiological investigation and physical examination suitable for the patient with characteristic symptoms of COVID-19 and, for confirmation, RT - qPCR test for coronavirus was performed in accordance with the Brazilian Ministry of Health guideline for COVID-19 management8. The study included all patients who tested RT - qPCR for coronavirus and were at least 18 years old, during the period from 03/01/2020 to 05/09/2020. The study was approved by the ethics committee of the Federal University of Espírito Santo (approval number: 4.120.872). Informed consent was obtained.
Data generation and reliability
The data relating to suspected and/or confirmed cases of COVID-19 were reported by nurses and doctors from the care units in the city of Vitória, in a notification form in the patient's electronic medical record. It is important to note that filling the notification is mandatory and failure to complete the notification makes it impossible to proceed with the filling of the patient's medical record.
The test used to diagnose the disease was the SARS-CoV-2 Antibody Test, Wondfo, China, with a sensitivity of 86,43%, a specificity of 99,57%, a positive predictive value of 99,68%, and a negative predictive value of 17,31%.
Predictors and outcomes
We used the following signs and symptoms as predictive variables: dyspnea, fever, nasal flap, intercostal circulation, cyanosis, cough, O2 saturation <95%, runny nose, odynophagia, diarrhea, nausea, vomiting, headache, adynamia, irritability, conjunctivitis, and convulsions. Besides that, comorbidities such as pulmonary, cardiovascular, renal, hepatic, diabetes, HIV infection, smoking, history of bariatric surgery, use of immunosuppressants, cancer, and chronic neurological issues, were utilized as predictive variables. Demographic variables such as gender, age in years, pregnancy, and travel history were also used. Predictor variables were selected through studies that described the symptoms and comorbidities prevalent in patients infected with SARS-CoV-29,10. The selected outcome was the diagnosis of COVID-19. Missing data were handled using K-Nearest Neighbors Imputer (KNNImputer/Scikit-Learn).
Statistical Analysis
The sample was described in absolute and relative values of the prevalence of symptoms and comorbidities between the different groups. To calculate the Odds Ratio, a multiple logistic regression model was performed using the Python Statsmodels library. Their values are described together with the respective p-value and confidence interval.
Machine Learning models applied
The predictive performances of seven machine learning models (Extra Tree Classifier, XGBoost, Random Forest Classifier, MLPClassifier, Gradient Boosting Classifier, Logistic Regression, and Support Vector Machine) were tested, obtained from the Scikit-Learn and XGBoost libraries applied in Python. For applying data to the models, the study sample was divided into training and testing to evaluate the model on a test set in the proportions of 70% and 30% respectively, using the train_test_split tool available in Scikit-Learn. The threshold defined for each algorithm was established in order to obtain better results for a screening test (high sensitivity and high negative predictive value).
To balance the training dataset we used SMOTE Tomek, a tool available from imbalanced-learn library which makes a process of over and undersampling without generating noisy samples from the oversampling of outliers11. To evaluate the performance of the classifying algorithms, we used subsequent metrics: area under the ROC curve (AUC), sensitivity, specificity, positive predictive value, and negative predictive value. Data reliability and machine learning model development were made according to transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD)12.
Explainability of the model
To identify which variables most impacted the model, to bring greater reliability and have a better explanation of its predictive power, the following tools were used: feature_importances_ (Scikit-Learn), SHAP (SHapley Additive exPlanations) and Drop Column Feature Importances.
RESULTS
The study sample was made of 888 COVID-19 positive patients and 1821 COVID-19 negative patients. After rebalancing the data, the training sample had 1263 patients with SARS-CoV-2 infection and 1410 patients negative for the disease. The test set maintained the proportion of the original sample with 170 patients with COVID-19 and 356 without.
The descriptive and statistical analysis of the continuous and categorical variables used as predictors within the entire sample of the study, in patients diagnosed with COVID-19 and in patients with excluded COVID-19 are shown in table 1 and table 2, respectively. Being a healthcare professional had a prevalence odds ratio (POR) of 1.48 associated with the presence of COVID-19. The symptoms of fever, cough, nasal congestion and cyanosis were associated with COVID-19 with the respective POR 2.14, 1.92, 1.58, and 4.07. The presence of diabetes and male gender were also associated with a greater chance of presenting COVID-19, with a POR of 1.80 and 1.27, respectively.
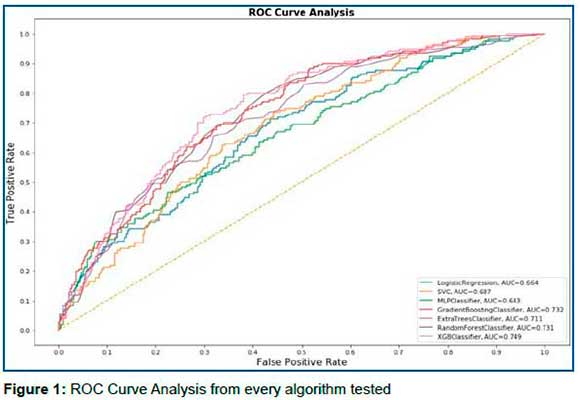
The algorithm with the best result was the XGBoost (XGB) which obtained an area under the ROC curve of 0.752, a sensitivity of 90%, specificity of 40%, positive predictive value (PPV) of 42.16%, and a negative predictive value (NPV) of 91.02%, in the test sample setting the threshold to 0.15. The result of each algorithm is contained in Table 3. Figure 1 contains the ROC curve for each algorithm.
The most important variables for the model's performance according to the feature importance of Scikit-Learn were fever, cough, history of international travel less than 14 days ago, male gender, and nasal congestion, respectively. In comparison, the SHapley tool's most important variables were fever, male gender, being a healthcare professional, cough, and history of international travel for less than 14 days, respectively.
DISCUSSION
Machine learning algorithms can positively impact the clinical decision of doctors13,14. The model proposed in the present study could be used as a screening tool for those who will need tests in order to rationalize the use of RT-qPCR tests to detect genetic material from the new coronavirus and to facilitate preventive actions regarding isolation.
Compared to the rapid test used for screening by PMV (Wondfo SARS-CoV-2 antibody test), which has a sensitivity of 86.43% and specificity of 99.57%, our model showed a superior result in terms of sensitivity (90%) and NPV (91.02%), however at the expense of lower specificity (40%). Some previously published models using variables from laboratory tests to diagnose COVID-19, such as those by Meng et al. obtained an area under the ROC curve (AUROC) of 0.872, a PPV of 86.35%, and a NPV of 84.62%5. Batista et al. using other laboratory variables contained in blood count and C-reactive protein obtained an AUROC of 0.847, sensibility 0.677, specificity 0.850, PPV 0.778, and NPV 0.77315. Although, none of it achieves better parameters for population triage (high sensibility and high NPV) neither proposes alternatives for a screening instrument. We consider that the clinical presentation of COVID-19, which has been presenting with nonspecific symptoms with many differential diagnoses of viral infections related to flu syndrome, can be related to the lower specificity of the model. Since patients with flu-like symptoms are required to test for COVID-19 as well, even in a scenario with endemic influenza circulation, where that patient might be infected with influenza or SARS-CoV-2, our model would still rise the pre-test probability. This can be explainable once the predictions would help exclude the patients that do not have the probability of COVID-19 and then the healthcare facility would just test him for Influenza, and not both Influenza and SARS-CoV-2.
As with the rapid test, our model is easier to apply than the other actual algorithms using laboratory variables, since, in the real scenario, especially in places with no integrated electronic medical records, there is extreme difficulty in integrating the data of medical records with laboratory results. The model proposed has a really low cost for its use, being able to be accessed by any device via the web platform and reaching the internet, while the rapid test has a cost of BRL 250.00 (~USD 45.00).
In addition, given that one of the major current discussions on the use of complex machine learning algorithmic models is "black box" behavior. This means that, even with excellent performance and potential in healthcare, some models can't be explained in a way in which parameters were used to arrive at the predicted16 and this fact becomes a struggle when using such models for medical decisions.
Therefore, we used the SHapley Additive exPlanation Values, present in the SHAP library, to explain the machine learning algorithms performed. In summary, SHapley, utilizes the game theory, assigning a value of importance to each variable present within the model's prediction17, thus, it is possible to map which were the most important variables for the outcome, consequently, it has a positive impact on the reliability of machine learning algorithms. The explainability of the most important features to the predictive power of a model can also contribute to further studies in which variables we should focus on to create scores, describe the disease and even interfere in treatment proposals.
The study has limitations in the generalization of its results, once the algorithms were trained in a population of a single city. There is also the possibility of Berkson's bias since the sample is composed only of people who sought care at a health facility. In addition, there was no differentiation and no analysis of other diseases with respiratory manifestations in the group of patients who received the diagnosis of "non COVID-19", which may explain the negative odds ratio for dyspnea and the diagnosis of COVID-19. It would be interesting to have blood oxygen saturation data, as this result could reflect on a phenomenon known as "happy hypoxia", which has been described in the current pandemic situation18.
The machine learning algorithms have limitations as a screening tool for asymptomatic patients, but the main goal of the study was to use them as a screening tool for the patients that would require the RT-PCR confirmation in healthcare facilities. In a scenario where you don't have many tests available to use, it is better to raise the pre-test likelihood of your patient than test everyone. Our models could be used to give directions in which patients would be tested (the positive ones in the model) and which wouldn't (the negatives one in the model).
However, our model was designed to predict the symptomatic patients, and even the cheaper serology laboratory tests looking for IgM and IgG anti-SARS-CoV-2, have low positive predictive value during the early symptomatic phase, being a good option only after around 7 days of its first symptoms.
CONCLUSION
The model performed high sensitivity and high NPV values, important characteristics for a screening tool, and has a very low cost for its uses since only clinical variables are needed for its decision making. The use of machine learning models as population triage could impact public health expenses in the face of the pandemic, rationalizing uses of diagnosis tests like RT-PCR or rapid test by serology.
Acknowledgments
We would like to thank the City Hall of Vitória and its Health Department for sharing the data and support this research.
Authors contributions
Gustavo Carreiro Pinasco: study design, data interpretation, writing, and review. Eduardo Moreno Júdice de Mattos Farina: study design, data interpretation, writing, literature search; Fabiano Novaes Barcellos Filho: data analysis, data interpretation, writing. Willer França Fiorotti: data interpretation and writing, literature search; Matheus Coradini: data analysis, data interpretation, building the machine learning model, figures; Sheila Cristina de Souza Cruz: data collection and writing. Andre Louzada Colodette: data analysis and data interpretation. Luciene Rossati Loureiro: data collection; Tatiane Comério: data collection; Dilzilene Cunha Sivirino Farias: data collection. Katia Valéria Manhabusque: writing and review. Eliane de Fátima Almeida Lima: data collection.
Competing interest statement
We declare no competing interest.
Data and code availability
The anonymized data and the code used to build the machine learning models are available at https://github.com/matheuscoradini/ml-covid-vix.
REFERENCES
1.WHO Coronavirus Disease (COVID-19) Dashboard. World Health Organization - Avaliable from: <https://covid19.who.int> (2020). [ Links ]
2.Bustin, S. & Nolan, T. RT-qPCR Testing of SARS-CoV-2: A Primer. International Journal of Molecular Sciences 21, 3004 (2020). DOI: https://doi.org/10.3390/ijms21083004 [ Links ]
3.Wynants, L. et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ 369, m1328 (2020). DOI: https://doi.org/10.1136/bmj.m1328 [ Links ]
4.Peiffer-Smadja, N., Maatoug, R., Lescure, FX. et al. Machine Learning for COVID-19 needs global collaboration and data-sharing. Nat Mach Intell 2, 293-294 (2020). DOI: https://doi.org/10.1038/s42256-020-0181-6 [ Links ]
5.Meng, Z. et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv, (2020). DOI: https://doi.org/10.1101/2020.03.18.20035816 [ Links ]
6.Yan, L. et al. A machine learning-based model for survival prediction in patients with severe COVID-19 infection. medRxiv, (2020). DOI: https://doi.org/10.1101/2020.02.27.20028027 [ Links ]
7.Zangirolami-Raimundo, J., Echeimberg, J. & Leone, C. Research methodology topics: Cross-sectional studies. Journal of Human Growth and Development 28, 356-360 (2018).DOI: https://doi.org/10.7322/jhgd.152198 [ Links ]
8.Orientações para o Manejo de Pacientes de COVID-19. Federal Government of Brazil (2020). Preprint at: <https://www.gov.br/saude/pt-br> [ Links ].
9.Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. & Napoli, R. Features, Evaluation, and Treatment of Coronavirus. (StatPearls Publishing LLC., 2020). [ Links ]
10.McIntosh, K., Hirsch, M. & Bloom, A. Coronavirus disease 2019 (COVID-19): Epidemiology, virology, and prevention. Uptodate (2020). Preprint at <https://www.uptodate.com/contents/coronavirus-disease-2019-covid-19-epidemiology-virology-clinical-features-diagnosis-and-prevention#H3103904400> [ Links ]
11.Batista, G., Prati, R. & Monard, M. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explorations Newsletter 6, 20-29 (2004). DOI: https://doi.org/10.1145/1007730.1007735 [ Links ]
12.Moons, K. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): Explanation and Elaboration. Annals of Internal Medicine 162, W1-W73 (2015).DOI: https://doi.org/10.7326/M14-0698 [ Links ]
13.Shah, P., Kendall, F., Khozin, S. et al. Artificial intelligence and machine learning in clinical development: a translational perspective. npj Digit. Med. 2, 69 (2019). DOI: https://doi.org/10.1038/s41746-019-0148-3 [ Links ]
14.Finding a role for AI in the pandemic. Nat Mach Intell 2, 291 (2020). DOI: https://doi.org/10.1038/s42256-020-0196-z [ Links ]
15.Batista, A., Miraglia, J., Donato, T. & Chiavegatto Filho, A. COVID-19 diagnosis prediction in emergency care patients: a machine learning approach. medRxiv (2020). DOI: https://doi.org/10.1101/2020.04.04.20052092 [ Links ]
16.Ribeiro, M., Singh, S. & Guestrin, C. "Why Should I Trust You?": Explaining the Predictions of Any Classifier. Association for Computing Machinery 1135-1144 (2016). DOI: https://doi.org/10.1145/2939672.2939778 [ Links ]
17.Lundberg, S. & Lee, S. A Unified Approach to Interpreting Model Predictions. 31st Conference on Neural Information Processing Systems NIPS (2017). Preprint at <https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html> [ Links ]
18.Couzin-Frankel, J. The mystery of the pandemic's 'happy hypoxia'. Science 368, 455-456 (2020). DOI: https://doi.org/10.1126/science.368.6490.455 [ Links ]
 Correspondence:
Correspondence:
Gustavo Carreiro Pinasco
gustavo.pinasco@ufes.br
Manuscript received: may 2021
Manuscript accepted: december 2021
Version of record online: june 2022